The NeuroGenomics and Informatics (NGI) Center focuses on deep molecular characterization of neurodegeneration and diseases of the central nervous system (CNS), with emphasis on Alzheimer disease (AD), Parkinson’s disease (PD), frontotemporal dementia (FTD), stroke. The ultimate goal of the center is to use high-throughput, multi-omic molecular profiling to understand the biology of these diseases, to define novel biomarkers of disease, and to identify novel therapeutic targets.
NeuroGenomics and Informatic Structure
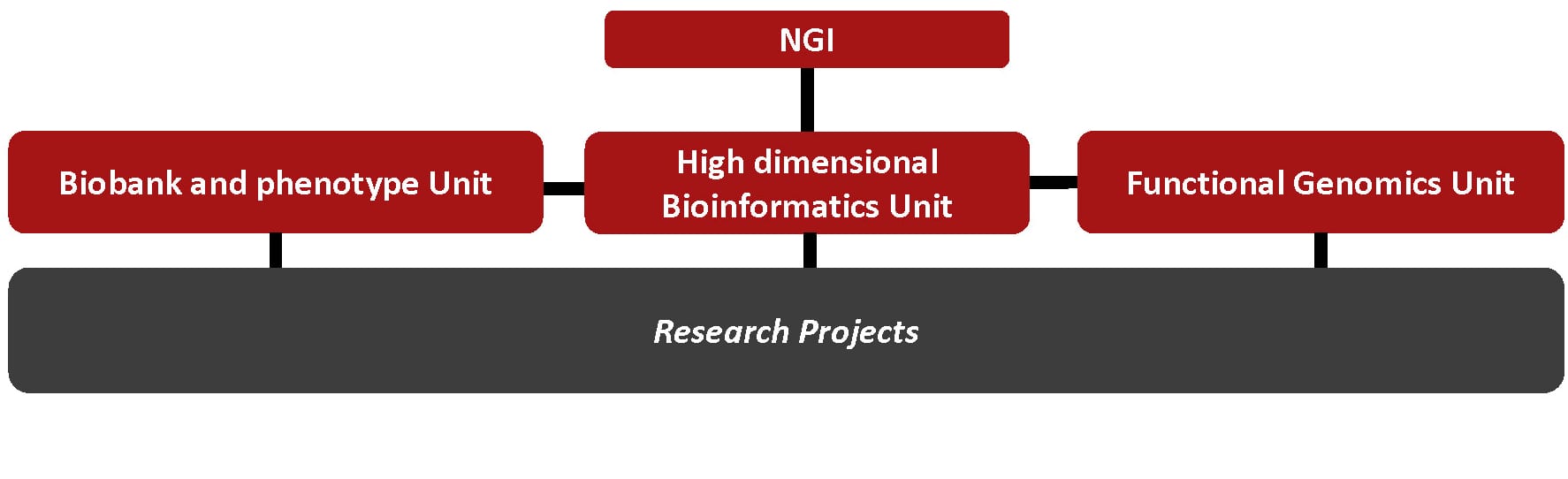 The NGI includes three research units that provide support, expertise, and data for specific projects.
The NGI includes three research units that provide support, expertise, and data for specific projects.
The Biobank and Phenotype Unit, harmonizes and combines available samples for AD, PD, FTD, stroke, and dystonia among other diseases and conditions. This unit receives samples from new participants, as well as longitudinal participants for other ongoing studies. In collaboration with the High-Dimensional Bioinformatics Unit, the Biobank and Phenotype Unit and the PIs of the specific projects select and prepare samples for data generation using GWAS, WES, WGS, RNA-seq, single nuclei RNA-seq, proteomics, epigenomics, metabolomics, and lipidomics.
The High-Dimensional Bioinformatics Unit, receives, processes, QCs, and analyzes all of the -omic data. The High-Dimensional Bioinformatics Unit will use standard pipelines for all the samples to guarantee that data generate across project is comparable and can be combine for additional transversals analyses.
The Functional Genomics Unit, use novel approaches such as iPSC, mouse models, and genome editing to functionally characterize novel genes and pathways identified by the projects performed under the NGI umbrella other studies.
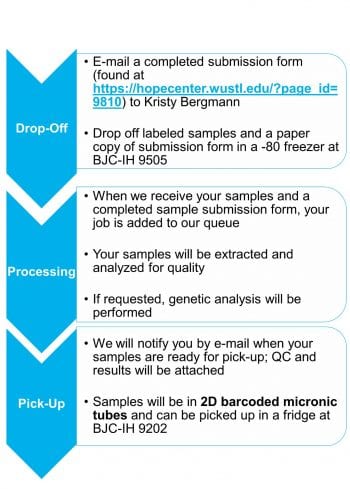
We partner with the Hope Center DNA and RNA Purification Core to obtain genetic material from research participants.
The Hope Center DNA and RNA Purification Core uses state-of-the-art instruments to extract high-quality DNA and RNA from multiple biological tissues.
The FlexSTAR+ instrument is used to obtain high molecular weight nuclear DNA from large volumes (1-10ml) of blood, buffy, buffy coat, or saliva.
The Maxwell 48 is used to extract DNA and RNA from low volumes (<1ml) of blood, buffy, Paxgene, bloodcards, and tissue as well as circulating DNA and RNA from plasma or cerebrospinal fluid.
DNA and RNA quality and quantity is assessed with the Agilent TapeStation 4200.
All samples are tracked using a 2D barcode system.
The Hope Center DNA and RNA Purification Core also offers other services such as DNA fingerprinting, Sanger Sequencing, APOE genotyping, custom genotyping of 1-30 SNPs, DNA normalization, and plating and biospecimen storage.
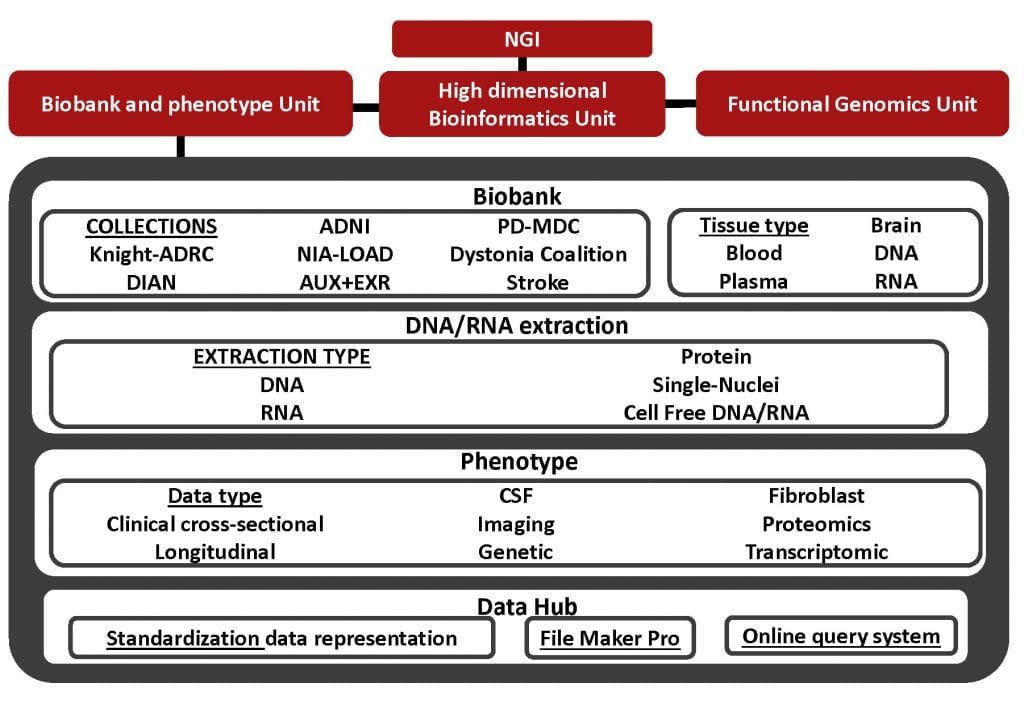
Unprecedented advances in sequencing and high-throughput -omic technologies have greatly increased the power and precision of analytical tools used in genomic research and accelerated the drive toward personalized medicine. Human biospecimens analyzed using these new and developing technology platforms have emerged as a critical resource for basic and translational research of neurodegenerative diseases. They are a direct source of molecular data from which targets for prevention, detection, and therapy are identified. The reliability of the data derived from these new platforms is dependent on the quality and consistency of the biospecimens being analyzed. As a result of the increased requirement for biospecimen quality, standardization of biospecimen resources using state-of the-art scientific approaches has become a pressing need across the research enterprise.
The goal of the Biobank and Phenotype Unit is to harmonize currently available biobanks and to guarantee the collection, processing, and distribution of authorized individual-level -omic information of samples from healthy individuals and those with neurodegenerative diseases. These samples have been characterized using uniform protocols including clinical assessment, psychometric testing, neuroimaging, and biomarker measurement as part of longitudinal studies of dementia and movement disorders. It is anticipated that collection of these data will facilitate clinical and basic science investigations of the pathogenesis of Alzheimer disease (AD), Parkinson’s disease (PD), frontotemporal dementia (FTD), tremor, and stroke among others.
To achieve these goals, the Biobank and Phenotype Unit will:
- Consolidate already available DNA collections in a single DNA bank and obtain GWAS and WGS/WES data for additional participants from Knight-ADRC, the Memory Diagnosis Clinic (MDC) at WU, the Movement Disorder Clinic participants, Stroke, and the Dystonia coalition and maintain the database of stored biological materials.
- We are consolidating our current DNA collections (Knight-ADRC, DIAN, ADNI, NIA-LOAD, MDC, Movement Disorder Clinic, Stroke and Dystonia Coalition) in a single DNA bank. All the DNAs will be storage in Barcode tubes and the concentration, quality, and location of the DNA will be storage in a database that will be developed in collaboration with the High dimensional Bioinformatics Unit.
- We continue collecting and properly storage blood samples, buffy coats, plasma, saliva and other biological material (bank the DNA, RNA, plasma and serum samples). We obtain skin biopsies from Knight-ADRC, MDC participants and Movement Disorder Clinic and bank the fibroblasts for studies using induced pluripotent stem cells.
- We extract DNA and RNA from blood and post-mortem brain using standardized state-of-the art technology to maintain sample integrity. We perform quality control (QC) assessment to evaluate the concentration and quality for each sample. Only samples that pass stringent QC step are sent for library prep and sequencing.
- In coordination with High Dimensional Bioinformatics Unit, we integrate our genetic data with those from other WUSTL cores for longitudinal clinical and clinic-pathological correlative analyses. We will review all pedigree information collected and expand family histories as appropriate.
- Generates genetic and multiomic (transcriptomic, epigenomic, proteomic, lipidomics and metabolomics) on selected deposit the data into the High Dimensional Bioinformatics Unit database.
- We DNA fingerprint all the samples each sample. As part of our quality control (QC) procedures we perform pairwise identity-by-state (IBS) distance and sex discordance analyses per sample.
- We screen for mutations in the most important AD, PD, and FTD genes in all the samples including APOE genotypes.
- We coordinate with Genome Technology Access Center (GTAC) and McDonnell Genome Institute for obtaining GWAS+exome chip data.
- We prepare and send the DNA for the selected participants from Projects 1, 2 and 3 to the McDonnell Genome Institute for WES or WGS sequencing.
- We validate sequencing data by using both medium and high-throughput genotyping technology and Sanger sequencing technology.
- Develops, curate and maintain a database of phenotypic and omic data.
- Collects phenotypic data. We use an already created database to record and storage all the important phenotypes for a specific disease/traits
- Curates and maintain the phenotypic data. We update, validate, curate the longitudinal clinical and clinicopathological data product of initial and periodical evaluations of our participants.
- Provides a centralized, confidential and secure access to phenotypic data for the cores and projects of the program as well as additional researchers and collaborators.
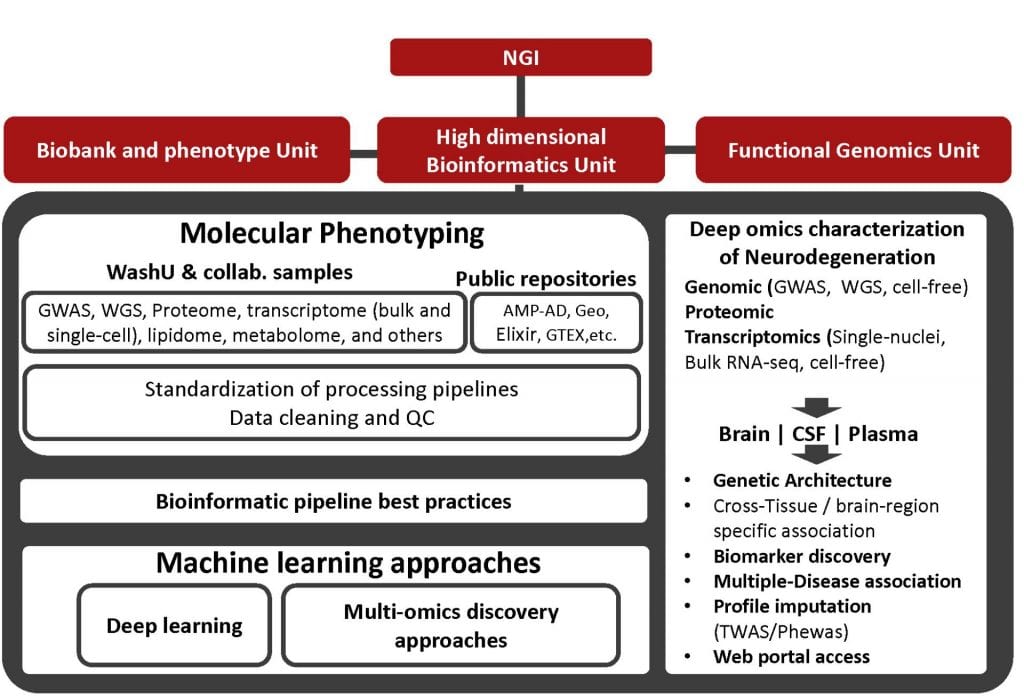
High-throughput technologies have been instrumental in generating a detailed molecular landscape of neurodegenerative diseases. However, the interpretation of such large amounts of data remains a challenge. This research unit is focused on the interrogation of large phenotypic, genomic, transcriptomic, proteomic and other omics datasets of neurodegenerative diseases to identify molecular profiles that provide novel insight into altered pathways associated with disease etiology and progression. We employ bioinformatic and machine learning approaches to aggregate distinct omics datasets to identify the genetic architecture of complex neurodegenerative diseases. High Dimensional Bioinformatics Unit we will process the high-throughput omic data generated for a large collection of brains, cerebrospinal fluid and plasma and brain single-cell sequencing data and coupling this data with genome-wide genotyping (GWAS and Whole Genome Sequencing) to generate complex hypothesis that can be later experimentally validated. The ultimate objective is: i) to increase our understanding of the etiology of neurodegeneration; ii) to enable early diagnosis of neurodegeneration; iii) to reveal novel intermediate traits involved in neurodegeneration and other complex traits; iv) to provide strategies for cohort selection for testing therapeutic targets; and v) to pave the road to identify disease-modifying treatments for neurodegenerative diseases.
The ability to mine into huge amount of information contained in biomedical Big Data will advance our understanding of neurodegeneration. To generate and validate novel hypothesis and obtain significant and relevant results is critical to have the data gathered, cleaned and accessible to researchers in a standardized procedure. This data is generated from different sources to capture the different aspects of the heterogeneity and complexity that characterize AD, PD or FTD between others. This includes an exhaustive ascertainment of the phenotypic characteristics of participants, as well as generation of detailed omic data.
Already available data: We have generated and/or downloaded GWAS data for 22,956 individuals, 3,275 WES, 2,343 WGS, and RNA-seq for 4,234 human brain samples. We also have generated single-nuclei RNA-seq data for four samples and 40 more samples are in the pipeline. We are currently generated proteomic data for 1278 CSF, 632 plasma and 459 brain samples.
The goal of the High Dimensional Bioinformatics Unit is to provide access to high-quality data and robust analytical pipelines that allow the best generation of data-driven hypothesis and their further interpretation. To do so, these specific aims are proposed.
- To develop, curate and maintain a repository of genetic and multi-omic information of participants. We will generate a repository that will provide centralized access to all data generated by the projects of the program, as well as additional studies, for example, accessions publicly available data in dbGaP.
- To process and control for the quality of the genotypes called from Next Generation Sequencing (NGS) and genome-wide arrays. We will provide the infrastructure and processing pipelines to QC, impute GWAS data and align and call high quality variants from NGS short reads.
- To process, QC, and deliver analysis ready proteomic, transcriptomic or other omic data. . We will provide the infrastructure and processing pipelines to clean the proteomic and transcriptomic studies.
- To integrate omic data gathered in the projects in a common repository. We will aggregate all of the data generated into a common digital repository.
- To provide a centralized, confidential and secure access to omic data for the Units and projects
- To provide standardized methods and procedures to analyze omic and phenotypic data. We will deploy, ascertain and provide access to the alternative tools that construct models to ascertain the relationships between omic data and phenotypes and ascertain their statistical significance (linkage, segregation and association) at the different biological levels (single variant analysis, burden tests for gene level analysis, pathways analysis, and heritability). We will also provide standardized methods to access, test and integrate external repositories of omics data (e.g. transcriptomics, proteomic, etc.)
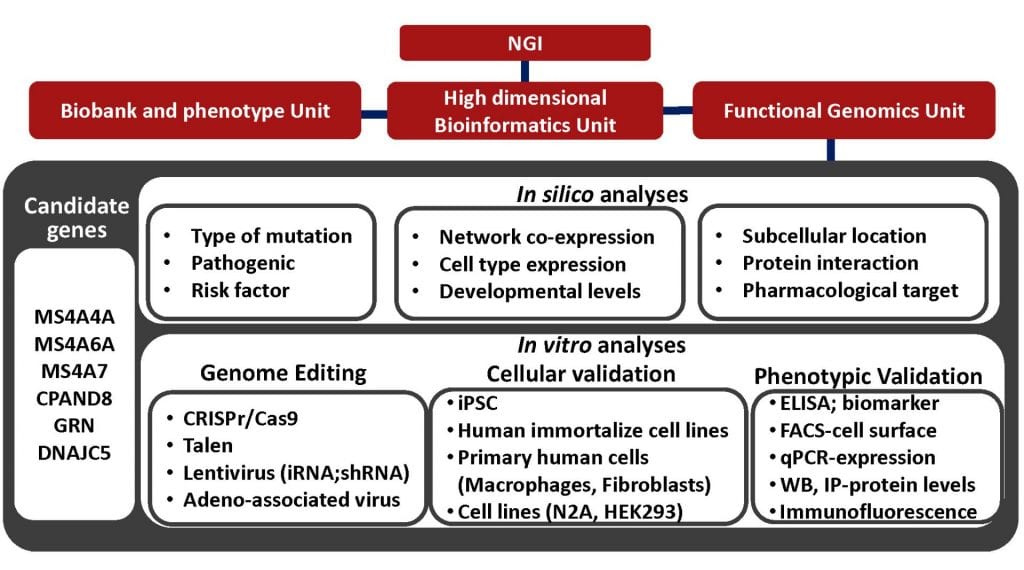
Currently, one of the main challenges in human molecular genetics is the interpretation of rare genetic variants of unknown clinical significance and the identification of “causal” genes in data from genome-wide association studies. The goal of the functional unit is to demonstrate the link between the variants/genes/pathways to reliable in vitro and in vivo models of neurodegenerative diseases. The functional genomic unit combines genome-wide association data, DNA/RNA sequencing data with gene-editing technologies, induced pluripotent stem cells (iPSc), and specific cellular phenotypes of neurodegenerative disease approaches for illuminating the molecular basis of these diseases. We use genome editing approaches to over-express, knock-down or knock-out entire genes, which will allow us to obtain information on the possible involvement of targeted genes in neurodegenerative pathways. We use RNA-Seq from iPSC-derived neurons, microglia and astrocytes obtained from primary fibroblasts from patients with pathogenic mutations in Mendelian genes or risk-associated variants with neurodegenerative diseases to understand the specific disruptions in gene networks in these cellular models. We use CRISPR-corrected isogenic cell lines as controls in which we modify the respective mutation or risk-associated variant. These data are analyzed together with “omics” data from CSF, brain and plasma from patients with neurodegenerative diseases. The knowledge of mechanisms through which gene variations modulate biological functions in normal and pathological conditions might open new avenues toward the design of novel and specific diagnostic, prophylactic or therapeutic interventions.
Translate the genetic and genomic findings to cell models is a necessary step to fully understand the role of specific genes in disease as well as to fully disentangle the biological processes that lead to neurodegeneration. Multiple large scale genetic and genomic studies including the Alzheimer’s Disease Sequencing Project, the Accelerating Medicines Partnership – Alzheimer’s Disease (AMP-AD), AMP-Parkinson Disease (AMP-PD), the UK biobank, and our own studies have and will identify novel variants, genes, networks and proteins implicated in neurodegeneration. Similarly to TREM2 or PLD3, it is expected that for some of those genes, very little will be known about the biology of these genes in healthy brains and in disease.
The main goal of the Functional Genomics Unit is to use cell models to:
1) determine the main role of the newly identified genes in health,
2) determine the functional mechanism by which the risk allele lead disease and
3) identify and test compounds that rescue the pathologic effect of the risk allele.
The Functional Genomics Unit has strong interactions with the High Dimensional Bioinformatics Unit and the Biobank and Phenotype Unit, as the High Dimensional Bioinformatics Unit identifies individuals that carry specific risk or protective variants, and the Biobank and Phenotype Unit obtains fibroblast from those specific individuals that later can be used for generate iPSC.